Edit detail for FrontPage revision 1 of 72
| 1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 28 29 30 31 32 33 34 35 36 37 38 39 40 41 42 43 44 45 46 47 48 49 50 51 52 53 54 55 56 57 58 59 60 61 62 63 64 65 66 67 68 69 70 71 72 | ||
|
Editor: damberger
Time: 2009/08/14 11:54:18 GMT+0 |
||
| Note: updated the news | ||
changed: - <img src="welcome.gif" /><br> <b>CARA is a program for assignment of NMR resonances in biomacromolecules and was developed in the group of <a href="http://www.mol.biol.ethz.ch/wuthrich/">Kurt Wüthrich Group</a> at the <a href="http://www.ethz.ch/">Swiss Federal Institute of Technology ETH</a> in Zurich, Switzerland. It continues to be developed and supported and can easily be installed on all major platforms</b> This is the front page of the <b>CARA documentation Wiki</b>. A <b>Wiki</b> is a platform for joint editing of HTML pages. We use the ZOPE version of a Wiki, called ZWiki. You'll find general Zwiki documentation at ZWiki:ZwikiDocs. The <b>CARA Wiki</b> has a HelpPage, RecentChanges, UserOptions and <a href="FrontPage/map">contents</a>. This Site is running on <a href="http://www.zope.org/">ZOPE</a> version 2.0. The Wiki can only be changed by members of the Cara Definition Team. We continue to welcome feedback and suggestions related to the Wiki and to Cara! Please send email regarding the wiki to the CDT. If you have a bug to report or feature you would like to see, you can use the "IssueTracker":Tracker. If you have problems which cannot be solved using the wiki, contact us. We like to hear from users! The <a href="mailto:info@nmr.ch">CDT</a> <h2>CARA Documentation</H2> - OverView<br> An overview of CARA's features. Here are some "credits":Credits. - "Download links":DownloadOverview <br> Links to download CARA, templates and documentation, as well as "CARA Release Information":Releases. Latest Alpha Release: **1.8.4a5** in April 2007 Latest Stable Release: **1.8.4** in April 2007 - GettingStarted <br> This is a brief introduction for new users explaining how to become productive with CARA. - "Tutorials":Tutorials <br> Many tutorials are available to introduce you to CARA's new tools and possibilities. - "**Frequently asked questions**":FAQ <br> - **Help and Support** <br> * **KnownBugs** and <a href="http://www.cara.ethz.ch/CARA/Wiki/CARACrashes">**CARACrash**</a> resources * <a href="http://www.cara.ethz.ch/CARA/Forum">**CaraForum**</a>: CARA's Support, Discussion and Announcement Forum * <a href="http://www.cara.ethz.ch/CARA/Tracker">**IssueTracker**</a>: Users should enter feedback on specific issues and bugs here. **NOTICE** Please enter the displayed numbers at the bottom to confirm an entry in the Tracker. Thanks! **NEWS**: "CorrectAtomMagnitudes.lua":CALUA has been added to the script collection. It sets all methyl atom magnitudes to 3 correcting an error in the standard template Start1.2.cara "WriteInputForMars.lua":CALUA has been added to the script collection. It writes out the chemical shift assignments for the systems in a format suitable for backbone assignment with MARS. "SparkyAssignmentsToCara.lua":CALUA has been added to the script collection. It imports the chemical shift assignments from a Sparky assignment list file into a CARA project. "WriteSparkyPeakList.lua":CALUA has been added to the script collection. It writes out a peaklist in sparky format from a CARA projects assignments. - **NEWS** Last update: August 14, 2009<br> <table> <td>"Latest News on CARA and Cara wiki":NewsPage.</td> </table> ** IN DEVELOPMENT** Extension of CARA model and interface to handle isotope-labeling. This will support the analysis of multiple samples in projects which contain two or more chains with different isotope labeling schemes, site-specific labeling, or partially labeled residues (e.g. perdeuteration with back-protonation). - **CALUA: CARA's scripting language** <br> <table> <td><img src="http://www.lua.org/images/powered-by-64.gif" /> </td> <td>"Documentation, Reference material and Supporting Scripts":CALUA for CARA's built-in scripting language based on LUA * >60 Scripts available as of Jan. 21, 2009.</td> </table> <HR> **Navigation in Cara wiki** You can return to this page at anytime by clicking on the Cara icon in the upper left corner of any CARA wiki page. You can go step-by-step through the wiki by clicking on the link next to "NEXT" at the bottom of each page. This will lead you through all aspects of Cara described in the wiki. Click on "The Basics":GettingStarted to get started. Click on "The Tutorials":Tools to skip the basics.
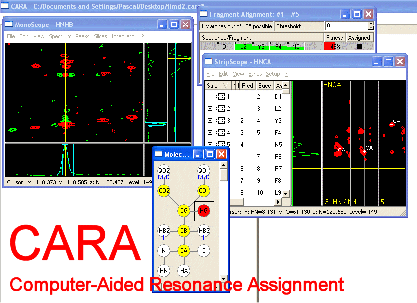
CARA is a program for assignment of NMR resonances in biomacromolecules and was developed in the group of Kurt Wüthrich Group at the Swiss Federal Institute of Technology ETH in Zurich, Switzerland. It continues to be developed and supported and can easily be installed on all major platforms
This is the front page of the CARA documentation Wiki. A Wiki is a platform for joint editing of HTML pages. We use the ZOPE version of a Wiki, called ZWiki?. You'll find general Zwiki documentation at ZWiki:ZwikiDocs. The CARA Wiki has a HelpPage?, RecentChanges?, UserOptions? and contents. This Site is running on ZOPE version 2.0.
The Wiki can only be changed by members of the Cara Definition Team. We continue to welcome feedback and suggestions related to the Wiki and to Cara! Please send email regarding the wiki to the CDT. If you have a bug to report or feature you would like to see, you can use the IssueTracker. If you have problems which cannot be solved using the wiki, contact us. We like to hear from users!
The CDT
CARA Documentation
- OverView?
An overview of CARA's features. Here are some credits. - Download links
Links to download CARA, templates and documentation, as well as CARA Release Information.Latest Alpha Release: 1.8.4a5 in April 2007
Latest Stable Release: 1.8.4 in April 2007
- GettingStarted?
This is a brief introduction for new users explaining how to become productive with CARA. - Tutorials
Many tutorials are available to introduce you to CARA's new tools and possibilities. - Frequently asked questions
- Help and Support
- KnownBugs? and CARACrash resources
- CaraForum: CARA's Support, Discussion and Announcement Forum
- IssueTracker: Users should enter feedback on specific issues and bugs here.
NOTICE Please enter the displayed numbers at the bottom to confirm an entry in the Tracker. Thanks!
NEWS:
CorrectAtomMagnitudes.lua has been added to the script collection. It sets all methyl atom magnitudes to 3 correcting an error in the standard template Start1.2.cara
WriteInputForMars.lua has been added to the script collection. It writes out the chemical shift assignments for the systems in a format suitable for backbone assignment with MARS.
SparkyAssignmentsToCara.lua has been added to the script collection. It imports the chemical shift assignments from a Sparky assignment list file into a CARA project.
WriteSparkyPeakList.lua has been added to the script collection. It writes out a peaklist in sparky format from a CARA projects assignments.
- NEWS Last update: August 14, 2009
Latest News on CARA and Cara wiki. IN DEVELOPMENT
Extension of CARA model and interface to handle isotope-labeling. This will support the analysis of multiple samples in projects which contain two or more chains with different isotope labeling schemes, site-specific labeling, or partially labeled residues (e.g. perdeuteration with back-protonation).
- CALUA: CARA's scripting language

Documentation, Reference material and Supporting Scripts for CARA's built-in scripting language based on LUA - >60 Scripts available as of Jan. 21, 2009.
Navigation in Cara wiki
You can return to this page at anytime by clicking on the Cara icon in the upper left corner of any CARA wiki page.
You can go step-by-step through the wiki by clicking on the link next to "NEXT" at the bottom of each page. This will lead you through all aspects of Cara described in the wiki.
Click on The Basics to get started.
Click on The Tutorials to skip the basics.