MoleculeViewer is a tool to create or modify ResidueTypes with a graphical interface. It is fairly intuitive to use. Click at the position where you would like to create or modify an atom and then use the context menu to perform the appropriate action. To link two atoms, click on the first one and then shift click on the second one. To remove the link repeat the same procedure. You can click-drag to move atoms.
Step-by-step procedure to create a ResidueType structure using MoleculeViewer.
You will need to know:
- covalent structure of the ResidueType
- names of each atom (e.g. IUPAC nomenclature)
- each atoms expected average chemical shift and deviation (4 x standard deviation)
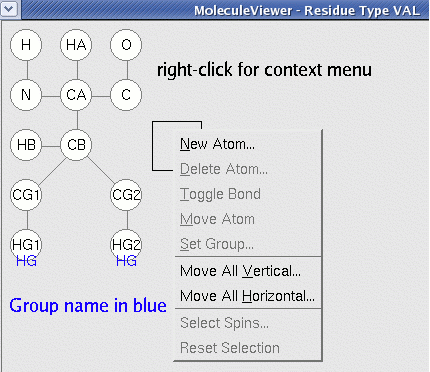
Steps
- Create atoms. Click at the appropriate location where you want to create an atom. The box marks the selected location on a 2D grid.
- Right-click "New Atom".
- Enter the Symbol which is the name of the atom (e.g. IUPAC name for ALA methyl is "HB").
- Enter the Magnitiude which is the number of equivalent spins with same shift (ALA methyl: 3).
- Select the Type which is the chemical identity (from periodic table) (ALA methyl: H).
- Group which is the name for the pseudoatom the atom belongs to (if several atoms have degenerate shift, the shift will be assigned this name).
- Mean is the average chemical shift observed for this atom.
- Deviation is the allowed deviation for the shift used in placing fragments into the sequence. In the standard template Start1.2.cara Deviation = 4 * Standard Deviation.
- Create a second atom and link it to the first.
- Click on first atom so that box appears around it.
- Shift-Click on second atom. A link appears between the two.
- To remove the link, repeat the procedure.
- Continue to create and link atoms until the whole structure is represented.
- Some useful tricks.
- You can move single atoms by click-dragging them.
- You can move the entire residue by using the context menu items "Move all vertical" or "Move all horizontal". NOTE: Do not move any atoms off the grid because CARA has a bug. If you do this accidentally, just repeat it using the same distance but opposite sign.
- You can create atoms that have no shift (like O) and link them in the ResidueType. They will have no affect on the Peak Inference.
- "Select Spins" is used to simulate the transfer of magnitization for a given step in an experiment procedure. Select the starting spin for the step and then "Select Spins" to see which spins are reached by this step.
Return to Creating ResidueType