The first step in homonuclear assignment is to pick correlations between amide and alpha protons in the fingerprint region of the COSY. Each amino acid gives rise to one fingerprint peak except for GLY which has two (H/HA2 & H/HA3) and PRO which has none.
Click on a peak in the FingerPrint region, and right-click selecting the context menu item pick new system. Alternatively, use the ShortCut py. Enter the labels "H" for x-axis, "HA" for y-axis, and click the remember CheckBox?. This will keep the current settings for the "Pick new system" dialog.
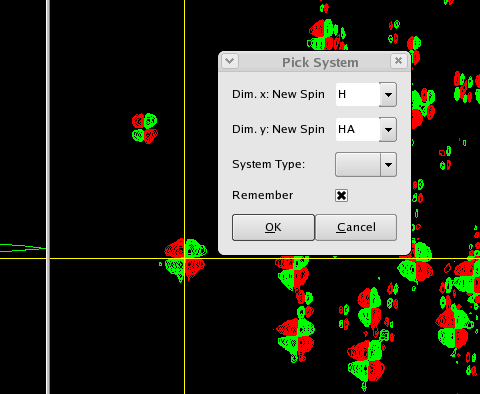
When you pick a fingerprint peak with "Pick new System", CARA creates a new spin-system and adds two spins, the amide proton "H" and the alpha proton "HA". The spin-system appears in the SystemList?. You can expand the system by clicking on it to see the two spins it contains. Note that the system is given a unique Id number (like every Object in the CARA object model). You cannot change this number, but you can assign the system to the correct Residue. CARA uses the spins "H" and "HA" to display three additional peaks:
The transposed crosspeak: x=HA, y=H (HA,H)
and
the two diagonal peaks: (H,H) and (HA,HA).
These peaks are generated dynamically from the SpinList? using PeakInference?. They are not independent objects which can be erased. If you move a peak, the spins shifts are changed and all other peaks involving these spins will move. This guarantees consistency. It is not possible to have inconsistent peak positions for two peaks involving the same spin.
It is important to ensure that spectra are correctly referenced so the diagonal is really the diagonal. See here for how to do this. It is also important to make sure that all spectra in the project are consistently referenced to one another. See here for how to do this.
Return to Homonuclear assignment