Edit detail for PolyScope revision 1 of 1
| 1 | ||
|
Editor: damberger
Time: 2007/01/24 15:22:08 GMT+0 |
||
| Note: | ||
changed: - PolyScope Introduction PolyScope is the logical extension of HomoScope into 3 dimensions. It predicts the position of peaks in displayed 3D spectra based on the list of assigned spins,the "SpectrumType":CreateSpectrumType definition, and the list of <a href="#Spinlinks">spin links</a>.<BR> Orientation of Spectra in Display If you open a 2D correlation spectrum like a 15N-HSQC with PolyScope, then you can select the 3D in the strip, and work exactly like you would do with SynchroScope. The advantage of PolyScope is that all the peaks are inferred based on the model and the choices for peak labels are determined automatically to match the SystemType. If you open a 3D with PolyScope: The 3D data is oriented such that the plane shows dimensions dim D1 and dim D2 (matching the HomoScope plane) and the Strip shows dim D3. E.g. 3D 15N-resolved ![1H,1H]-NOESY: <BR> Plane X=1H(acq) and Y=1H(noe) and Strip Y=15N. PolyScope as alternative to SynchroScope You can use PolyScope in exactly the same way as SynchroScope. E.g. Select an HSQC15N with PolyScope. Now select the 3D spectrum to display in the strips. You can control which spins are combined to display peaks in the plane using the "Plane" menu items: * Show SpinLinks (to show peaks connecting two linked spins) * Show Inferred Peaks (connects spins within a residue which are expected to give rise to peaks. e.g. HN-N in HSQC15N) * Show Unlabeled Peaks (connects unlabeled spins to all other spins in residue. e.g. all "?" (atomtype H) to all "?" (atomtype N) ) * Show Neighbor Spins (include neighbor spins like "CA-1" in peak inference e.g for HNCA) Setting up Parameters to Display 3D GhostPeaks (Peak width and Peak Depth) Real 3D crosspeaks are not points in space. They extend in 3 dimensions in the 3D spectrum. Consequently, the intensity from a given peak is observed for a range of ppm above and below the plane where it is centered. CARA represents the physical size of peaks as an adjustable parameter, *peak width*, one parameter for each dimension of a 3D experiment. This accounts for the fact that each dimension has a typical linewidth for the peaks (due to the limited sampling and the window function). CARA uses the peakwidths to decide whether a crosspeak symbol "+" is displayed for a given peak. Each peak is displayed only if the peak position is within the peakwidth (in ppm) of the currently displayed planes ppm. You should also determine the peak width in the horizontal dimension of the Strips. Therefore in a 3D, each dimension has a characteristic *peakwidth* (in ppm). The width of the strips (in ppm) is determined automatically by CARA by multiplying the *peakwidth* for the strip by the *width factor*. The width factor can be set interactively anytime in StripScope or PolyScope using the command *wf* followed by a real number. A fairly useful value is wf=2. This ensures that the strips are wide enough to show the edges of the peaks in the system as well as any nearby peaks. Tools available Because PolyScope is derived from HomoScope, it has all the useful tools for extending assignments that are available in HomoScope, such as *Propose Spin...* in the Strip window, and *Propose Peak...* in the Plane window. PolyScope can be used to confirm the existence of expected peaks in 3D spectra, and to extend the assignments into spin systems where conventional means have failed. Examples The following image shows the appearance of a 3D 15N-resolved ![1H,1H]-NOESY opened with PolyScope. The 2D plane is shown with all expected spin pairs between the two 1H dimensions independent of their 15N chemical shift. (Note that <a href="#Spinlinks">spin links</a> were generated from a 3D peaklist of this NOESY) <img src="polyscope1.gif" /><br> After selecting a spin pair in the plane, the strips and 1D slice <BR> are shown with the Y-axis of the strip displaying the 15N dimension<BR> of the 3D at the anchor position of the spin pair chosen in the plane. * Select a spin in the strip. * Use the option *Plane-Show 3D Plane* to show the ![1H,1H]-plane at<BR> the 15N chemical shift of this spin. This allows the user to inspect<BR> the 3D spectrum ![1H,1H] plane by plane. <img src="polyscope2.gif" /><br> <a name="Spinlinks">Spin links</a> SpinLinks are Caras way of representing through-space connectivity<BR> of the NOESY spectrum. I.e. if a longrange NOE exists between two<BR> 1H spins, then a spinlink can be created to represent this fact.<BR> Cara will display crosspeaks corresponding to this NOE in all<BR> NOESY spectra where they are expected automatically.<BR> SpinLinks are automatically created when you use *propose peak*<BR> to pick a peak in a NOESY plane. You can control the display<BR> of peaks derived from Spinlinks using the option *View-Show Spin Links*<BR> SpinLinks are represented in the !SystemList as a node attached to<BR> each spin. The symbol for a spinlink is two balls connected by<BR> a line. E.g. if you use *propose peak* to create a spinlink between HN of<BR> system 1 (spin 1) and HA of system 3 (spin 10) you will see the<BR> peak in each NOESY spectrum where you expect it. In addition in<BR> the SystemList you will find a spinlink in system 1 from the HN (spin 1)<BR> to the HA of system 3 (spin 10) and in system 3 from the HA to HN of<BR> system 1 (spin 1). You can also create spinlinks from an XEASY project so that the NOEs<BR> are visible in your Cara project. How to define Spinlinks from XEASY project: Requirements: You must have defined the Spins using a Proton List from the XEASY project.<BR> (*Import AtomList* from Cara Explorer). You must have a NOESY spectrum<BR> with the corresponding peaklist representing the NOEs. 1. Select the NOESY spectrum from the main CARA window. <BR> 2. Open the spectrum with MonoScope. <BR> 3. Import a Peaklist using *Peaks-Import Peaklist*. <BR> 4. Generate the SpinLinks using *Peaks-Import !SpinLinks* <BR> Using the standard options will hide the SpinLinks in spectra other <BR> than the one where you imported the SpinLinks to.<BR> Every pair of assigned 1H spins occuring in a line of the peaklist will<BR> generate a spinlink between the two spins. <BR> E.g. to ensure that PolyScope displays all the spins in the <BR> NOESY towers of a 3D 15N-resolved ![1H,1H]-NOESY,<BR> like in the examples given here, you can import the<BR> peaklist from the 3D 15N-resolved NOESY into MonoScope and then <BR> generate the SpinLinks. See also the example PolyScopeRotated <BR> "The Scopes":Tools BACK: HomoScope NEXT: PolyScopeRotated
PolyScope
Introduction
PolyScope is the logical extension of HomoScope into 3 dimensions. It predicts the position of peaks in displayed 3D spectra based on the list of assigned spins,the SpectrumType definition, and the list of spin links.
Orientation of Spectra in Display
If you open a 2D correlation spectrum like a 15N-HSQC with PolyScope, then you can select the 3D in the strip, and work exactly like you would do with SynchroScope. The advantage of PolyScope is that all the peaks are inferred based on the model and the choices for peak labels are determined automatically to match the SystemType?.
If you open a 3D with PolyScope:
The 3D data is oriented such that the plane shows dimensions dim D1 and dim D2 (matching the HomoScope plane) and the Strip shows dim D3.
E.g. 3D 15N-resolved [1H,1H]-NOESY:
Plane X=1H(acq) and Y=1H(noe) and Strip Y=15N.
PolyScope as alternative to SynchroScope
You can use PolyScope in exactly the same way as SynchroScope. E.g. Select an HSQC15N with PolyScope. Now select the 3D spectrum to display in the strips. You can control which spins are combined to display peaks in the plane using the "Plane" menu items:
- Show SpinLinks (to show peaks connecting two linked spins)
- Show Inferred Peaks (connects spins within a residue which are expected to give rise to peaks. e.g. HN-N in HSQC15N)
- Show Unlabeled Peaks (connects unlabeled spins to all other spins in residue. e.g. all "?" (atomtype H) to all "?" (atomtype N) )
- Show Neighbor Spins (include neighbor spins like "CA-1" in peak inference e.g for HNCA)
Setting up Parameters to Display 3D GhostPeaks (Peak width and Peak Depth)
Real 3D crosspeaks are not points in space. They extend in 3 dimensions in the 3D spectrum. Consequently, the intensity from a given peak is observed for a range of ppm above and below the plane where it is centered. CARA represents the physical size of peaks as an adjustable parameter, peak width, one parameter for each dimension of a 3D experiment. This accounts for the fact that each dimension has a typical linewidth for the peaks (due to the limited sampling and the window function). CARA uses the peakwidths to decide whether a crosspeak symbol "+" is displayed for a given peak. Each peak is displayed only if the peak position is within the peakwidth (in ppm) of the currently displayed planes ppm.
You should also determine the peak width in the horizontal dimension of the Strips. Therefore in a 3D, each dimension has a characteristic peakwidth (in ppm). The width of the strips (in ppm) is determined automatically by CARA by multiplying the peakwidth for the strip by the width factor. The width factor can be set interactively anytime in StripScope or PolyScope using the command wf followed by a real number. A fairly useful value is wf=2. This ensures that the strips are wide enough to show the edges of the peaks in the system as well as any nearby peaks.
Tools available
Because PolyScope is derived from HomoScope, it has all the useful tools for extending assignments that are available in HomoScope, such as Propose Spin... in the Strip window, and Propose Peak... in the Plane window.
PolyScope can be used to confirm the existence of expected peaks in 3D spectra, and to extend the assignments into spin systems where conventional means have failed.
Examples
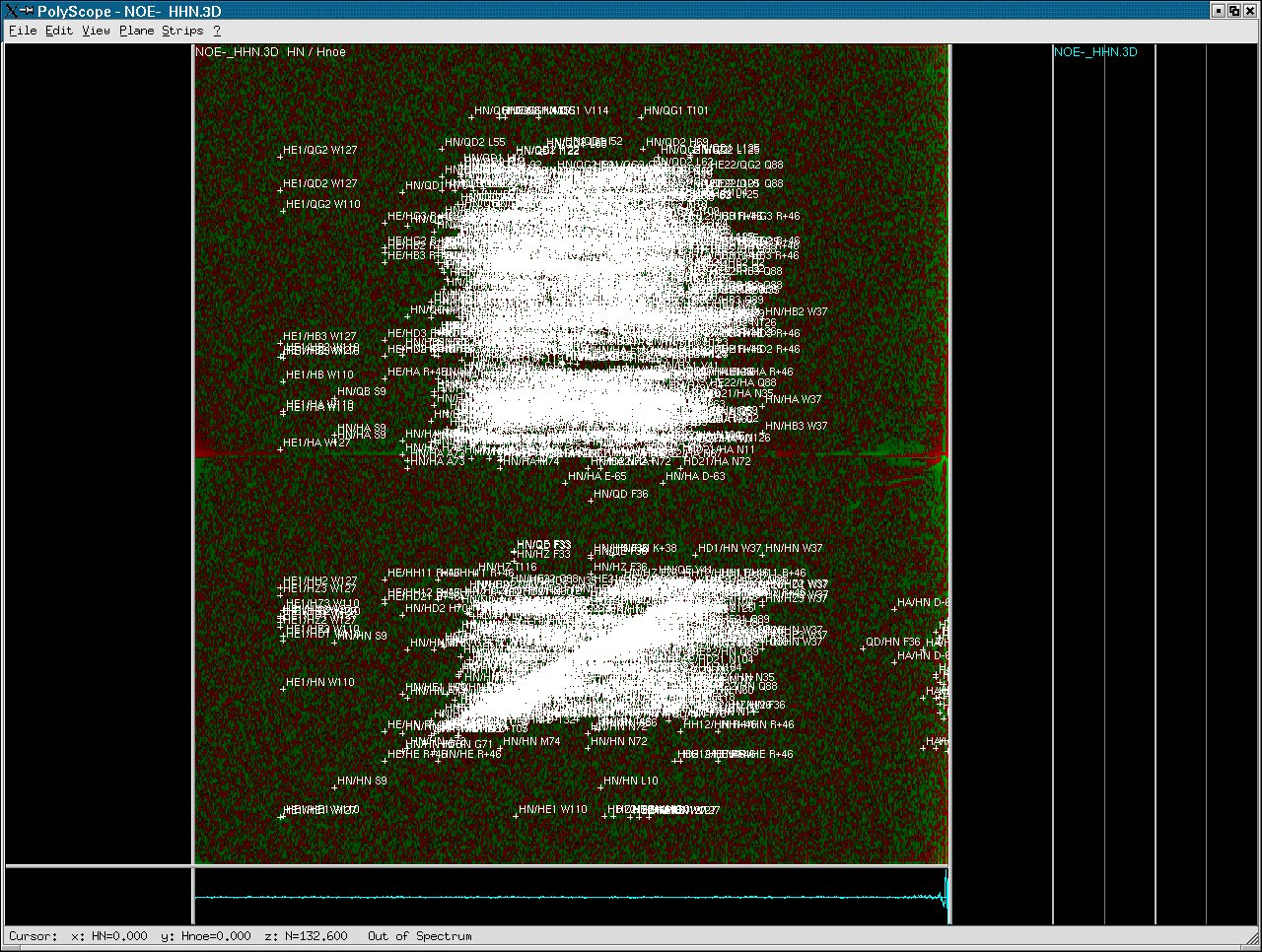
The following image shows the appearance of a 3D 15N-resolved [1H,1H]-NOESY opened with PolyScope. The 2D plane is shown with all expected spin pairs between the two 1H dimensions independent of their 15N chemical shift. (Note that spin links were generated from a 3D peaklist of this NOESY)
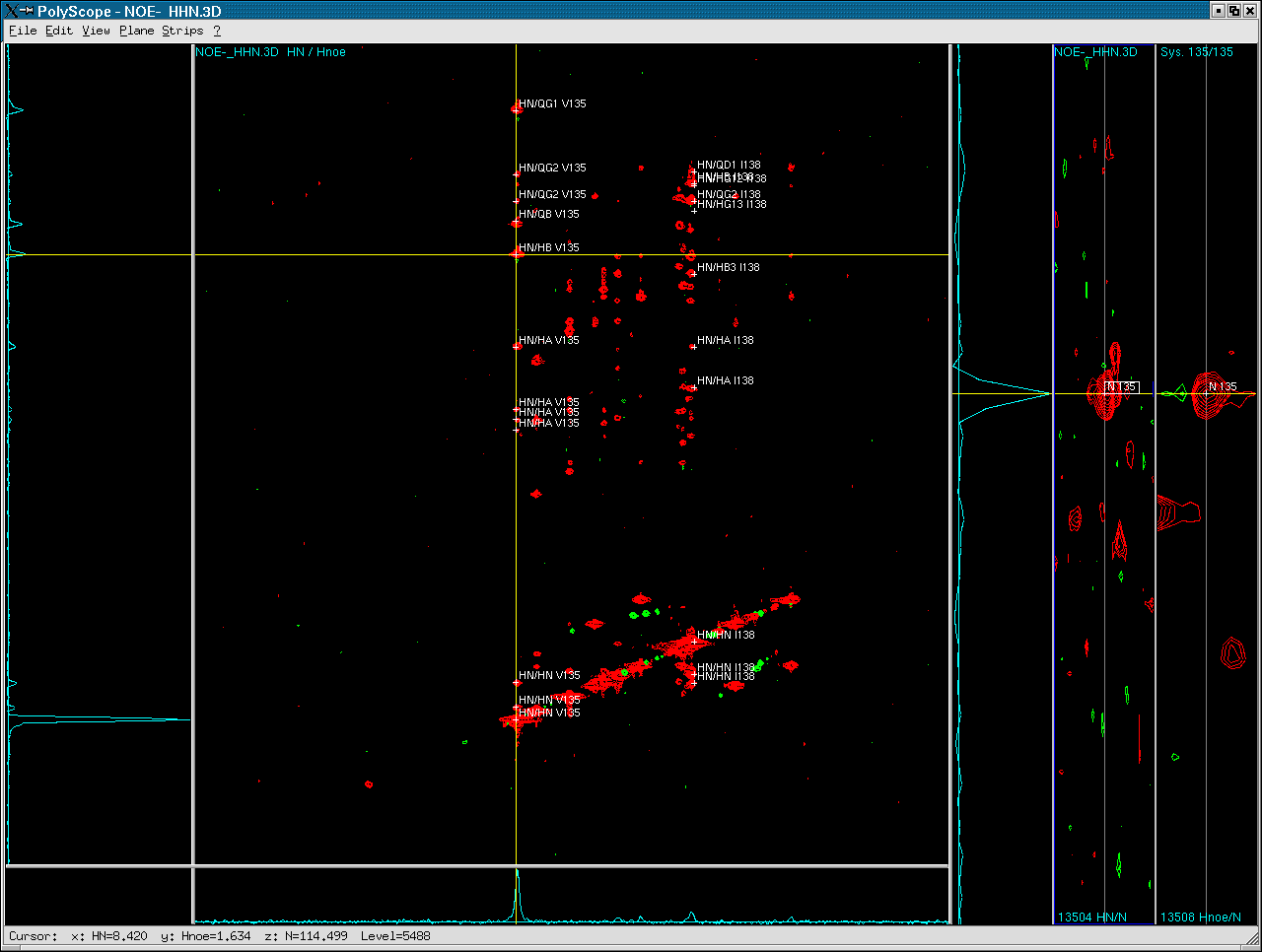
After selecting a spin pair in the plane, the strips and 1D slice
are shown with the Y-axis of the strip displaying the 15N dimension
of the 3D at the anchor position of the spin pair chosen in the plane.
- Select a spin in the strip.
- Use the option Plane-Show 3D Plane to show the [1H,1H]-plane at
the 15N chemical shift of this spin. This allows the user to inspect
the 3D spectrum [1H,1H] plane by plane.
Spin links
SpinLinks are Caras way of representing through-space connectivity
of the NOESY spectrum. I.e. if a longrange NOE exists between two
1H spins, then a spinlink can be created to represent this fact.
Cara will display crosspeaks corresponding to this NOE in all
NOESY spectra where they are expected automatically.
SpinLinks are automatically created when you use propose peak
to pick a peak in a NOESY plane. You can control the display
of peaks derived from Spinlinks using the option View-Show Spin Links
SpinLinks are represented in the SystemList as a node attached to
each spin. The symbol for a spinlink is two balls connected by
a line.
E.g. if you use propose peak to create a spinlink between HN of
system 1 (spin 1) and HA of system 3 (spin 10) you will see the
peak in each NOESY spectrum where you expect it. In addition in
the SystemList? you will find a spinlink in system 1 from the HN (spin 1)
to the HA of system 3 (spin 10) and in system 3 from the HA to HN of
system 1 (spin 1).
You can also create spinlinks from an XEASY project so that the NOEs?
are visible in your Cara project.
How to define Spinlinks from XEASY project:
Requirements:
You must have defined the Spins using a Proton List from the XEASY project.
(Import AtomList from Cara Explorer). You must have a NOESY spectrum
with the corresponding peaklist representing the NOEs?.
- Select the NOESY spectrum from the main CARA window.
- Open the spectrum with MonoScope.
- Import a Peaklist using Peaks-Import Peaklist.
- Generate the SpinLinks using Peaks-Import SpinLinks
Using the standard options will hide the SpinLinks in spectra other
than the one where you imported the SpinLinks to.
Every pair of assigned 1H spins occuring in a line of the peaklist will
generate a spinlink between the two spins.
E.g. to ensure that PolyScope displays all the spins in the
NOESY towers of a 3D 15N-resolved [1H,1H]-NOESY,
like in the examples given here, you can import the
peaklist from the 3D 15N-resolved NOESY into MonoScope and then
generate the SpinLinks. See also the example PolyScopeRotated
BACK: HomoScope
NEXT: PolyScopeRotated