Edit detail for StripScope revision 1 of 1
| 1 | ||
|
Editor: damberger
Time: 2006/04/04 16:51:35 GMT+0 |
||
| Note: links updated after rename | ||
changed: - StripScope StripScope is CARA's tool for heteronuclear backbone assignment and sequence mapping. It can display strips from 3D spectra and link then together and map the resulting fragments onto the sequence. <img src="stripscope1.gif" /><br> How to use it? 1. We will assume, that all backbone spectra were picked with SynchroScope. <BR><BR> 2. Select any strip in the list to the left. * Use *Show All Successors* to display the best candidates for the i+1 neighbor to the selected strip. The first strip is the reference strip which was selected. The following strips are ordered reading from left to right starting with the best candidate. The ranking is obtained according to chemical shift matching (e.g CA-1 of reference with CA of candidate). * Use *Show All Predecessors* to display the best candidate strips for the i-1 neighbor to the reference strip (e.g. CA of reference matches CA-1 spin of candidate). * Use *Show Free Successors* and *Show Free Predecessors* to display only unlinked candidate strips (see below) * It is advisable to use *Show All ...* to be sure to always find the best matches. <BR><BR> 3. Use CARA's tools to decide on the correct successor/predecessor. * Compare the slices by clicking on a candidate. The slice for the reference strip (the leftmost strip) is shown in cyan. The slice for the currently active strip is shown in green. * Use *View > Select Ruler Residue..* to display the expected ranges for CA, CB and other chemical shifts for any amino acid in the sequence. * Use *Assignment Candidates...* to reduce the choice of residues (e.g. turn off Glycines or Prolines in HNCA; turn on/off appropriate residues for spectra of residue-specific selectively labeled samples. When you find a good match between the reference strip and the active strip, Link them by using *Link to Reference*. The link will be entered into the Striplist. See the columns *Pred* and *Succ*. If you change your mind and want to erase a link select one of the strips that are linked and use *Unlink Successor* or *Unlink Predecessor*. Once several strips are linked together you will have a fragment.<BR><BR> 4. Local Mapping onto the sequence is a very powerful tool. Any fragment can be mapped onto the sequence and assigned. * Select any strip in the fragment and select *Show Alignment...*. The *Show alignment* window will show you all possible alignments. The alignments are ranked according to the quality of the chemical shift matching to the statistical database in the residue library. Quality of the chemical shift match for individual residues in an alignment are indicated by the green bars. If your fragment length exceeds 4 amino acids, there is often only one plausible mapping. Select the best fragment and use the context menu in the Fragment Alignment Window to *Assign* the selected fragment. Assigned Strips appear in the column labeled *Ass* in the Striplist. <img src="stripscope2.gif" /><br> You can export strips and anchor lists, as well as output files for MAPPER, a tool for global mapping. More details are <a href="http://www.cara.ethz.ch/CARA/Wiki/WorkingWithOtherProgramsImportExport#export">here</a>. "The Scopes":Tools BACK: SynchroScope NEXT: SystemScope
StripScope
StripScope is CARA's tool for heteronuclear backbone assignment and sequence mapping. It can display strips from 3D spectra and link then together and map the resulting fragments onto the sequence.
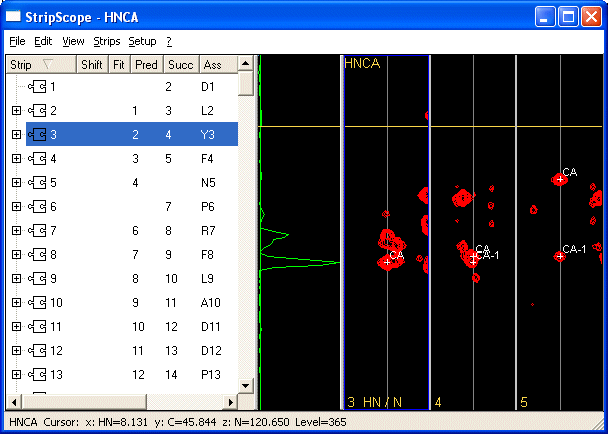
How to use it?
- We will assume, that all backbone spectra were picked with SynchroScope.
- Select any strip in the list to the left.
- Use Show All Successors to display the best candidates for the i+1 neighbor to the selected strip. The first strip is the reference strip which was selected. The following strips are ordered reading from left to right starting with the best candidate. The ranking is obtained according to chemical shift matching (e.g CA-1 of reference with CA of candidate).
- Use Show All Predecessors to display the best candidate strips for the i-1 neighbor to the reference strip (e.g. CA of reference matches CA-1 spin of candidate).
- Use Show Free Successors and Show Free Predecessors to display only unlinked candidate strips (see below)
- It is advisable to use Show All ... to be sure to always find the best matches.
- Use CARA's tools to decide on the correct successor/predecessor.
- Compare the slices by clicking on a candidate. The slice for the reference strip (the leftmost strip) is shown in cyan. The slice for the currently active strip is shown in green.
- Use View > Select Ruler Residue.. to display the expected ranges for CA, CB and other chemical shifts for any amino acid in the sequence.
- Use Assignment Candidates... to reduce the choice of residues (e.g. turn off Glycines or Prolines in HNCA; turn on/off appropriate residues for spectra of residue-specific selectively labeled samples.
When you find a good match between the reference strip and the active strip, Link them by using Link to Reference. The link will be entered into the Striplist. See the columns Pred and Succ. If you change your mind and want to erase a link select one of the strips that are linked and use Unlink Successor or Unlink Predecessor. Once several strips are linked together you will have a fragment.
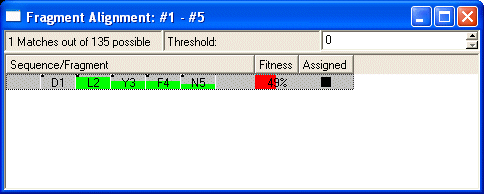
- Local Mapping onto the sequence is a very powerful tool. Any fragment can be mapped onto the sequence and assigned.
- Select any strip in the fragment and select Show Alignment.... The Show alignment window will show you all possible alignments. The alignments are ranked according to the quality of the chemical shift matching to the statistical database in the residue library. Quality of the chemical shift match for individual residues in an alignment are indicated by the green bars. If your fragment length exceeds 4 amino acids, there is often only one plausible mapping. Select the best fragment and use the context menu in the Fragment Alignment Window to Assign the selected fragment. Assigned Strips appear in the column labeled Ass in the Striplist.
You can export strips and anchor lists, as well as output files for MAPPER, a tool for global mapping. More details are here.
BACK: SynchroScope
NEXT: SystemScope