Edit detail for SynchroScope revision 1 of 1
| 1 | ||
|
Editor: damberger
Time: 2007/08/31 16:19:54 GMT+0 |
||
| Note: | ||
changed: - SynchroScope SynchroScope allows simultaneous manipulation of 2D and 3D-spectra. <img src="synchroscope.gif" /><br> How to use it? 1. Display a 2D spectrum with SynchroScope in the plane. Three empty strips appear to the right.<BR><BR> 2. Right-clicking the middle or right strip and selecting *Select Spectrum* in the resulting context menu shows a list of 3D spectra with compatible dimensions (meaning that two dimensions from the 2D and 3D have matching unique labels e.g. HN and N).<BR><BR> 3. The three strips display data from the 3D and are all centered at the x- and y- axis positions of the cursor in the Plane. Vertical axis of strip = y-axis of the 3D (e.g. CA dimension of 3D HNCA) * *Left* 1D trace. * *Center* 2D strip. Strip x-axis = x-axis of 3D. (E.g. HN dimension of 3D HNCA). * *Right* 2D strip. Strip x-axis = z-axis of 3D. (E.g. N dimension of 3D HNCA).<BR> Center lines appear along the y-axis of the strips to indicate the chemical shift of the cursor in the plane. 3. To calibrate a 3D vs a 2D take the following steps: * Place the cursor on a signal in the plane of the 2D and *Pick System*. "Screenshot":PickSystem * Pick a strong resolved peak in the Strip of the 3D "Screenshot":PickLabelIn3D * use "*Plane >Show 3D Plane*":ShowThreeD to view the plane of the 3D spectrum in place of the 2D.<BR> The "depth" of the plane is determined by the cursor position along the vertical axis in the Strip. You should see a strong signal near the position of your System in the plane from the 3D. * Move the cursor onto the "signal":SynchScopeCenterCrossHair. * select the crosspeak of the system using "*shift-click*":ShiftClickSystem. * Execute the command *Plane > "Calibrate from System*":CalibrateFromSystem.<BR> Now the 3D will be displayed shifted by the offset between your system in the 2D and its position in the 3D. "Second Screenshot":CalibrateFromSystem You can see the value of this offset calibration if you click on Spectra in the Main CARA window, and then click on the spectrum name. This will expand the node for the spectrum and show all relevant data for it including calibration offsets. 4. Alternatively, use the projection of the 3D corresponding to the 2D to calibrate the 3D (for this purpose, use PolyScope): * Calculate the projection of the 3D corresponding to the two dimensions of the 2D using the CARA-explorer "Tool-Project" (e.g. for HNCA, calculate the HN-N projection: HN-N_proj_of_HNCA) * Load the Projected experiment into the Project (e.g. HN-N_proj_of_HNCA.nmr) * Open the HSQC15N with PolyScope and place the cursor on a peak and *Pick New System* followed by OK. You can do this for several peaks if you wish. * In the Plane menu, Select Spectrum-Projected 3D (e.g. HN-N_proj_of_HNCA.nmr) * Click on the new System, move the cursor to its position in the projected 3D, and select "Plane-Calibrate" * In the CARA Explorer: Click on Spectra. Click on the puzzle piece for the projected Spectrum and the 3D to expand the views so you can see the parameters of the dimensions for both spectra. * Right-click on the 3D and select "Calibrate Spectrum". Enter the "cal" values appearing in the two dimensions of the projected 3D into the corresponding fields of the calibrate spectrum dialog: e.g. For the HNCA, you would copy cal value of "H(H) D1" from the HN-N_proj_of_HNCA.nmr into "Dim X(D1)" of the HNCA and cal value of "(N)N D2" from the HN-N_proj_of_HNCA.nmr into "Dim Z(D3)" of the HNCA. 5. Now calibrate any other spectra you wish to use. For backbone assignment, you may want to calibrate HNCA, HNCACB, CBCA(CO)NH onto the ![1H,15N]-HSQC.<BR><BR> 6. You may need to calibrate the third dimensions of the 3Ds onto each other. You can pick a spin in the 3D dimension (strip y-axis) select it, select a second spectrum, adjust the horizontal cursor to the related peak in the displayed spectrum and use *Strips > Calibrate Strip*.<BR><BR> 7. The fastest way to pick spectra for a backbone assignment, is the following: * Pick a system in a 2D, pick the two spins in the HNCA (note that the 1D-trace usually allows you to identify the stronger intraresidue peak as CA and the weaker sequential peak as CA-1) * Change to the CBCA(CO)NH. This should enable you to decide which of the previously picked spins was CA-1 and which one was CA. You can immediately label both of them from the right-click *label spin* menu. Now pick and label the second spin in this spectrum which is CB-1. * Switch to the HNCACB and pick and label the final spin as CB. Then go to the next 2D system in the ![1H,15N]-HSQC and repeat from step a.<BR><BR> 8. You can load a 15N-resolved 3D-NOESY spectrum into SystemScope and use sequential HN-peaks to check assignments or resolve ambiguities.<BR><BR> 9. If one of your spectra shows markedly different chemical shifts from the others (different experimental conditions), use the *Move Spin Alias* function to adapt your peaks to this spectrum.<BR><BR> * move the cursor to the real position of the peak in the spectrum. * select the crosspeak by using "shift-click". * use right-click context menu *Move Spin Alias* to define the position of the spin for this spectrum. *Move Spin* will move your peak for all spectra. The procedure to *Move Spin* is identical to that for *Move Spin Alias*. "The Scopes":Tools BACK: SliceScope NEXT: StripScope
SynchroScope
SynchroScope allows simultaneous manipulation of 2D and 3D-spectra.
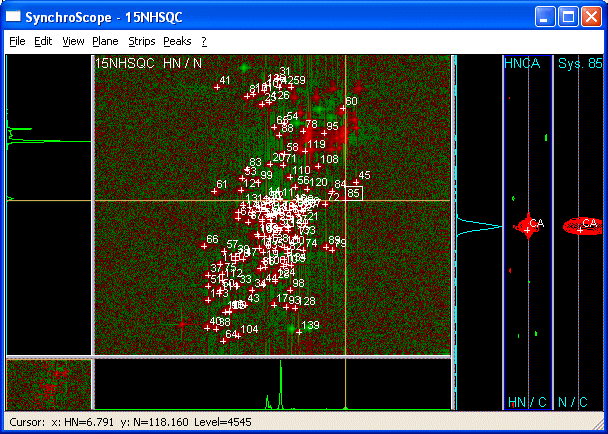
How to use it?
- Display a 2D spectrum with SynchroScope in the plane. Three empty strips appear to the right.
- Right-clicking the middle or right strip and selecting Select Spectrum in the resulting context menu shows a list of 3D spectra with compatible dimensions (meaning that two dimensions from the 2D and 3D have matching unique labels e.g. HN and N).
- The three strips display data from the 3D and are all centered at the x- and y- axis positions of the cursor in the Plane. Vertical axis of strip = y-axis of the 3D (e.g. CA dimension of 3D HNCA)
- Left 1D trace.
- Center 2D strip. Strip x-axis = x-axis of 3D. (E.g. HN dimension of 3D HNCA).
- Right 2D strip. Strip x-axis = z-axis of 3D. (E.g. N dimension of 3D HNCA).
Center lines appear along the y-axis of the strips to indicate the chemical shift of the cursor in the plane.
- To calibrate a 3D vs a 2D take the following steps:
- Place the cursor on a signal in the plane of the 2D and Pick System. Screenshot
- Pick a strong resolved peak in the Strip of the 3D Screenshot
- use Plane >Show 3D Plane to view the plane of the 3D spectrum in place of the 2D.
The "depth" of the plane is determined by the cursor position along the vertical axis in the Strip. You should see a strong signal near the position of your System in the plane from the 3D. - Move the cursor onto the signal.
- select the crosspeak of the system using shift-click.
- Execute the command *Plane > Calibrate from System*.
Now the 3D will be displayed shifted by the offset between your system in the 2D and its position in the 3D. Second Screenshot You can see the value of this offset calibration if you click on Spectra in the Main CARA window, and then click on the spectrum name. This will expand the node for the spectrum and show all relevant data for it including calibration offsets.
- Alternatively, use the projection of the 3D corresponding to the 2D to calibrate the 3D (for this purpose, use PolyScope):
- Calculate the projection of the 3D corresponding to the two dimensions of the 2D using the CARA-explorer "Tool-Project" (e.g. for HNCA, calculate the HN-N projection: HN-N_proj_of_HNCA)
- Load the Projected experiment into the Project (e.g. HN-N_proj_of_HNCA.nmr)
- Open the HSQC15N with PolyScope and place the cursor on a peak and Pick New System followed by OK. You can do this for several peaks if you wish.
- In the Plane menu, Select Spectrum-Projected 3D (e.g. HN-N_proj_of_HNCA.nmr)
- Click on the new System, move the cursor to its position in the projected 3D, and select "Plane-Calibrate"
- In the CARA Explorer: Click on Spectra. Click on the puzzle piece for the projected Spectrum and the 3D to expand the views so you can see the parameters of the dimensions for both spectra.
- Right-click on the 3D and select "Calibrate Spectrum". Enter the "cal" values appearing in the two dimensions of the projected 3D into the corresponding fields of the calibrate spectrum dialog:
e.g. For the HNCA, you would copy cal value of "H(H) D1" from the HN-N_proj_of_HNCA.nmr into "Dim X(D1)" of the HNCA and cal value of "(N)N D2" from the HN-N_proj_of_HNCA.nmr into "Dim Z(D3)" of the HNCA.
- Now calibrate any other spectra you wish to use. For backbone assignment, you may want to calibrate HNCA, HNCACB, CBCA(CO)NH onto the [1H,15N]-HSQC.
- You may need to calibrate the third dimensions of the 3Ds onto each other. You can pick a spin in the 3D dimension (strip y-axis) select it, select a second spectrum, adjust the horizontal cursor to the related peak in the displayed spectrum and use Strips > Calibrate Strip.
- The fastest way to pick spectra for a backbone assignment, is the following:
- Pick a system in a 2D, pick the two spins in the HNCA (note that the 1D-trace usually allows you to identify the stronger intraresidue peak as CA and the weaker sequential peak as CA-1)
- Change to the CBCA(CO)NH. This should enable you to decide which of the previously picked spins was CA-1 and which one was CA. You can immediately label both of them from the right-click label spin menu. Now pick and label the second spin in this spectrum which is CB-1.
- Switch to the HNCACB and pick and label the final spin as CB. Then go to the next 2D system in the [1H,15N]-HSQC and repeat from step a.
- You can load a 15N-resolved 3D-NOESY spectrum into SystemScope and use sequential HN-peaks to check assignments or resolve ambiguities.
- If one of your spectra shows markedly different chemical shifts from the others (different experimental conditions), use the Move Spin Alias function to adapt your peaks to this spectrum.
- move the cursor to the real position of the peak in the spectrum.
- select the crosspeak by using "shift-click".
- use right-click context menu Move Spin Alias to define the position of the spin for this spectrum.
Move Spin will move your peak for all spectra. The procedure to Move Spin is identical to that for Move Spin Alias.
BACK: SliceScope
NEXT: StripScope