Edit detail for SystemScope revision 1 of 1
| 1 | ||
|
Editor: damberger
Time: 2005/11/08 15:58:27 GMT+0 |
||
| Note: | ||
changed: - SystemScope SystemScope is CARA's sidechain assignment tool. It is called SystemScope because it is used primarily for building up spin-systems. The SystemScope window (show below) is divided into the following subwindows: a. *left dropdown menu*: displays sequential assignment for the currently selected spin-system if already defined b. *right dropdown menu*: allows user to temporarily assign a spin-system to a specific residue-type e.g. ALA c. *left top window*: spin list associated with the spin system, their chemical shift, and their assignment labels d. *left bottom window*: anchor list. Links between spins expected using the current "spectrum type":CreateSpectrumType along current X and Y axes) e.g. !HcCH-COSY links H with directly attached C along the X and Y axis - for VAL: HA-CA HB-CB QG1-CG1 QG2-CG2 e. *slice window*: displays slice from the center of the strip f. *strip window*: displays strip at the position of the currently selected anchor (e.g. shown QG2-CG2) from the list of anchors g. *plane window*: displays planes from the 3D - orientation and position of plane is set by user <img src="systemscope1.gif" /><br> How to use it? 1. We will assume, that all backbone spectra were picked with SynchroScope as discussed there. We will further assume that some fragments are already mapped onto the sequence using StripScope. 2. SystemScope is started by right-clicking onto any 3D-spectrum. It is possible to change between all spectra sharing the dimension types, e.g. !HcCH-COSY and 13C-resolved ![H,H]-NOESY. 3. Select any System using the left dropdown box. CARA will calculate which anchors are visible in this type of spectrum for this amino acid. If the anchors are strange, check your <a href="http://www.mol.biol.ethz.ch:8060/CARA/Wiki/CreateSpectrumType">Spectrum Type Definitions</a>, the mistake is most likely here. An expected anchor will also not appear if one or both spins are not yet assigned. 4. Double-Click on any anchor to display the third dimension of your spectrum. A slice appears to the left of it. Use *Show Depth...* to display the Plane passing through the diagonal peak. You can use this to adjust your anchor position. 5. Pick any off-diagonal peak. You can label it immediately, or leave it unknown. You can also apply a preliminary assignment to it using *Force Label...* Include the prefix ? in front of your assignment ( e.g. ?CG1 ). 6. Now select the new peak and choose *Show Orthogonal...* to find its own diagonal peak. <img src="systemscope2.gif" /><br> 7. Using this function you can pick and assign a Spin in the Orthogonal Plane and thus create a new anchor. Repeat this walk through the sidechain till it is fully assigned. Other Use Cases for SystemScope * No H-C anchors for a residue? - Assign some more spins. If you are unsure, use preliminary assignments with the ? prefix. - Combine the information of different spectra. If you have CA and CB, N and HN from triple resonance spectra, use a 15N-resolved TOCSY/NOESY, HNHA or HNHB to get H assignments. If you use TOCSY or NOESY, it may be unclear what to label the spins. Take a guess! CARA will use the newly assigned spins to generate new Anchors in the anchor list. Then you can try out different potential assignments. If your hunch is right, peaks should appear at the expected positions in the strips for all generated anchors. - Use HomoScope to pick spins in a 2D (e.g. ![1H,13C]-HSQC) * Fragment not mapped onto sequence Use the right pulldown-menu to temporarily assign a residue type to the system. CARA will calculate anchors assuming the selected residue-type. The residue-type and anchors will be forgotten as soon as you switch to another residue. * Unknown HSQC-peak Use HomoScope to pick and perhaps assign the new peak. It will get a new system ID. Then you can work with SystemScope to expand this new system. If you determine that this new peak is part of an existing system, use !HomoScopes *Eat System...* function to add it to the existing system. Select the destination system and then execute *Eat System...* You will be asked for the system number of the system to eat. * Rotated SystemScope Occasionally, you may want to look at a spectrum in a non-typical orientation. Example: You have assigned CA and CB of a system but not HA or HB. Open an (H)CCH-COSY using Rotated SystemScope. A pop-up window shows the standard orientation: <img src="systemscope3.gif" /><br> Change the orientation, so that the H dimension becomes the y-dimension (dimension 1): <img src="systemscope4.gif" /><br> A new SystemScope will appear with CA-CA, CA-CB, CB-CA and CB-CB as anchors. You can now select the anchors and assign the HA and HB spins in the strip window. "The Scopes":Tools BACK: StripScope NEXT: HomoScope
SystemScope
SystemScope is CARA's sidechain assignment tool. It is called SystemScope because it is used primarily for building up spin-systems. The SystemScope window (show below) is divided into the following subwindows:
a. left dropdown menu: displays sequential assignment for the currently selected spin-system if already defined
b. right dropdown menu: allows user to temporarily assign a spin-system to a specific residue-type e.g. ALA
c. left top window: spin list associated with the spin system, their chemical shift, and their assignment labels
d. left bottom window: anchor list. Links between spins expected using the current spectrum type along current X and Y axes)
e.g. HcCH-COSY links H with directly attached C along the X and Y axis - for VAL: HA-CA HB-CB QG1-CG1 QG2-CG2
e. slice window: displays slice from the center of the strip
f. strip window: displays strip at the position of the currently selected anchor (e.g. shown QG2-CG2) from the list of anchors
g. plane window: displays planes from the 3D - orientation and position of plane is set by user
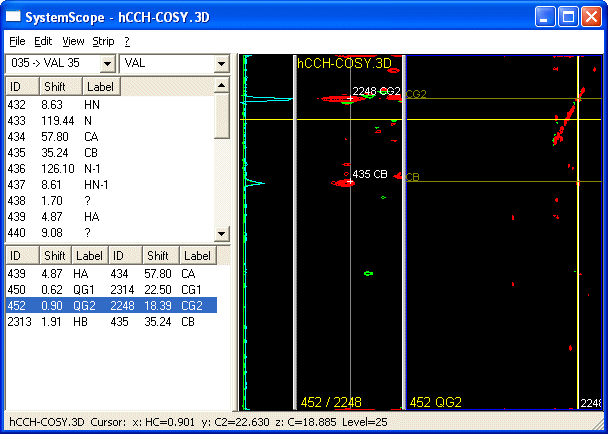
How to use it?
- We will assume, that all backbone spectra were picked with SynchroScope as discussed there. We will further assume that some fragments are already mapped onto the sequence using StripScope.
- SystemScope is started by right-clicking onto any 3D-spectrum. It is possible to change between all spectra sharing the dimension types, e.g. HcCH-COSY and 13C-resolved [H,H]-NOESY.
- Select any System using the left dropdown box. CARA will calculate which anchors are visible in this type of spectrum for this amino acid. If the anchors are strange, check your Spectrum Type Definitions, the mistake is most likely here. An expected anchor will also not appear if one or both spins are not yet assigned.
- Double-Click on any anchor to display the third dimension of your spectrum. A slice appears to the left of it. Use Show Depth... to display the Plane passing through the diagonal peak. You can use this to adjust your anchor position.
- Pick any off-diagonal peak. You can label it immediately, or leave it unknown. You can also apply a preliminary assignment to it using Force Label... Include the prefix ? in front of your assignment ( e.g. ?CG1 ).
- Now select the new peak and choose Show Orthogonal... to find its own diagonal peak.
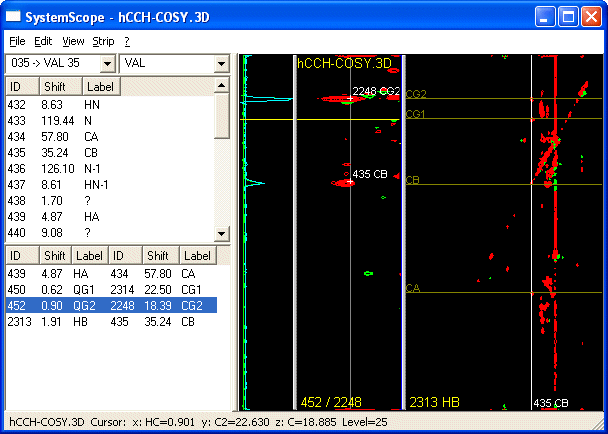
- Using this function you can pick and assign a Spin in the Orthogonal Plane and thus create a new anchor. Repeat this walk through the sidechain till it is fully assigned.
Other Use Cases for SystemScope
- No H-C anchors for a residue?
- Assign some more spins. If you are unsure, use preliminary assignments with the ? prefix.
- Combine the information of different spectra. If you have CA and CB, N and HN from triple resonance spectra, use a 15N-resolved TOCSY/NOESY, HNHA or HNHB to get H assignments. If you use TOCSY or NOESY, it may be unclear what to label the spins. Take a guess! CARA will use the newly assigned spins to generate new Anchors in the anchor list. Then you can try out different potential assignments. If your hunch is right, peaks should appear at the expected positions in the strips for all generated anchors.
- Use HomoScope to pick spins in a 2D (e.g. [1H,13C]-HSQC)
- Fragment not mapped onto sequence
Use the right pulldown-menu to temporarily assign a residue type to the system. CARA will calculate anchors assuming the selected residue-type. The residue-type and anchors will be forgotten as soon as you switch to another residue.
- Unknown HSQC-peak
Use HomoScope to pick and perhaps assign the new peak. It will get a new system ID. Then you can work with SystemScope to expand this new system. If you determine that this new peak is part of an existing system, use HomoScopes Eat System... function to add it to the existing system. Select the destination system and then execute Eat System... You will be asked for the system number of the system to eat.
- Rotated SystemScope
Occasionally, you may want to look at a spectrum in a non-typical orientation. Example: You have assigned CA and CB of a system but not HA or HB. Open an (H)CCH-COSY using Rotated SystemScope. A pop-up window shows the standard orientation:
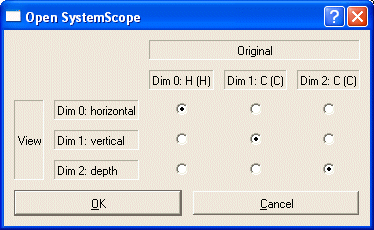
Change the orientation, so that the H dimension becomes the y-dimension (dimension 1):
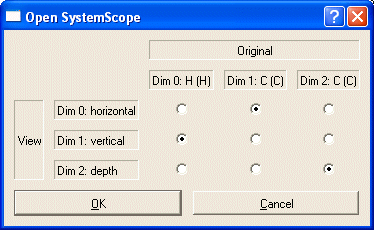
A new SystemScope will appear with CA-CA, CA-CB, CB-CA and CB-CB as anchors.
You can now select the anchors and assign the HA and HB spins in the strip window.
BACK: StripScope
NEXT: HomoScope